By Micki Maes
The segmentation of cortical and subcortical structures of the brain is required for numerous types of quantitative neuroanatomic evaluations. The volume and shape of brain structures, such as ventricles, hippocampus, and entorhinal cortex, are used to characterize disease states such as Alzheimer’s Disease.
Magnetic resonance imaging (MRI) provides detailed information of normal and pathologic anatomy and is a significant imaging modality in the diagnosis of brain disorders. Segmentation of cortical and subcortical structures from MR brain scans is a critical task that has many applications, including volumetric evaluations and shape analysis. Segmenting structures is also necessary in various types of clinical studies assessing morphological changes correlated with medical treatments. Even though manual segmenting by experts is still routine practice for high quality dissection, it is time-consuming, expensive, and subjective. Due to these drawbacks, automated segmentation tools have become a popular solution.
Most automated segmentation tools use a static atlas where the target image, likelihoods, and class priors are gathered over a large cohort of individuals. As such, individuals whose brain anatomy differs considerably from the target population will produce poor segmentation results due to the simple fact that the atlas information is not characteristic of the individual.
Cortechs.ai’s novel and innovative Dynamic Atlas (covariate modulated) technology is based on an impartial diffeomorphic atlas with probabilistic spatial priors built from a large training dataset of MR images. This multifactorial approach allows the use of the probabilistic atlas at age ranges where anatomical changes occur most rapidly; for instance, children and geriatric populations. This results in a more precise and dependable segmentation in all subjects, independent of age.
![]()
When applying the Dynamic Atlas for automatic structural segmentation, MR images are first corrected for head rotation and tilt, intensity inhomogeneity, skull stripped, and intensity calibrated to the atlas.
Creation of the Dynamic Atlas used age and gender as predictors (covariates). A large cohort of individuals undergoes NeuroQuant registration and segmentation using a Static Atlas. This yields a set of transformation parameters, voxel intensities, and segmentation parameters for each specific individual. The Dynamic Atlas approach involves using a statistical model to fit smooth functions of the predictor variables (age, sex covariates) to the output data of NeuroQuant segmentation. The resulting model parameters can then be used “on the fly” to “predict” the precise atlas that is optimal for segmentation of any individual.
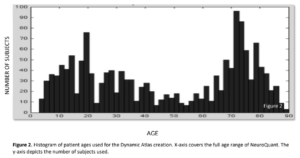
Data from tens of thousands of subjects were compiled to create the Dynamic Atlas. Data was collected on all three major scanner manufacturers: Siemens, GE, and Phillips at both 1.5T and 3.0T. The age range of individuals covered a broad age range from 3-100 years old, with particular attention to coverage of ranges where rates of volume change are the highest. Prior to fitting the model, data was inspected to ensure quality of initial segmentation and registration with atlas. Those subjects that did not meet the stringent quality criteria were discarded from further analysis.
The Dynamic Atlas framework also provides benefits over more advanced “multi-atlas” segmentation approaches by applying a continuous and computationally efficient statistical model as a prediction engine. The model is fit to the observed data, which includes both the original image data and derived data (e.g., registration and segmentation parameters), using smooth functions of the predictor variables. In this way, any combination of covariate values (age and gender) that makes up an individual can then be used to predict the atlas parameters that are appropriate for that individual. With the Dynamic Atlas, we generate a custom target image, class likelihoods, and class priors, in a principled model-based approach specific to each individual in a manner that cannot be achieved with static or multi-atlas approaches.
The main advantage of using the Dynamic Atlas versus the traditional static atlas or multi-atlas approach for image segmentation is that it allows a broader range of individuals to be segmented with greater accuracy and precision. The Dynamic Atlas approach to segmentation relies on prior information to be representative of the individual to be segmented. By incorporating flexible priors over a continuous range of demographic information, the Dynamic Atlas offers a principled and optimal approach to extend the segmentation capabilities of NeuroQuant over a broader range of individuals.
References:
- Rajapakse J.C.; Giedd J.N.; Rapoport J.L., (1997). Statistical approach to segmentation of single-channel cerebral MR images., IEEE Trans Med Imaging Apr; 16(2):176-86.
- Saeed N. (1998). Magnetic resonance image segmentation using pattern recognition, and applied to image registration and quantification., NMR Biomed Jun-Aug; 11(4-5):157-167.
- Clarke L.P. Velthuizen R.P., Camacho M.A., Heine J.J., Vaidyanathan M., Hall L.O., Thatcher R.W., & Silbiger M.L., (1995). MRI segmentation: methods and applications., Magn Reson Imaging; 13(3):343-368.
- Collins,D.L.,PetersT.M.,DaiW.,Evans,A.C.(1992).ModelbasedSegmentationofIndividualBrainStructuresfrom MRI Data. In Robb, R. A. (Ed.): Visualization in Biomedical Computing II, Proc. SPIE 1808. Chapel Hill, NC, 10-23.
- CollinsL.,EvansA.C.,HolmesC.,PetersT.M.(1995).Automatic3Dsegmentationofneuroanatomicalstructuresfrom MRI. Proceedings of Annual Conference on Information Processing in Medical Imaging (IPMI): 139-152.
- Zhang Y., Brady M., & Smith S. (2001). Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm., IEEE Trans Med Imaging Jan; 20(1):45-57.
- Held K., Kops E.R., Krause B.J., Wells W.M., Kikinis R., & Muller-Gartner H.W. (1997). Markov random field segmentation of brain MR images., IEEE Trans Med Imaging Dec; 16(6):878-86.
- VanLeemputK., MaesF.,VandermeulenD.,&SuetensP.(1999).Automatedmodel-basedtissueclassificationofMR images of the brain., IEEE Trans Med Imaging Oct; 18(10):897-908.
- CollinsL.,HolmesC.,PetersT.M.,&EvansA.C.(1995).Automatic3DModel- BasedNeuroanatomicalStructuresfrom MRI. Human Brain Mapping 3(3):190-208.
- Nowinski W.L., Fang A., Nguyen B.T., Raphel J.K., Jagannathan L., Raghavan R., Bryan R.N., & Miller G.A. (1997). Multiple brain atlas database and atlas-based neuroimaging system., Comput Aided Surg; 2(1):42-66.